Introduction to Magnetic Bead-Based Nucleic Acid Extraction
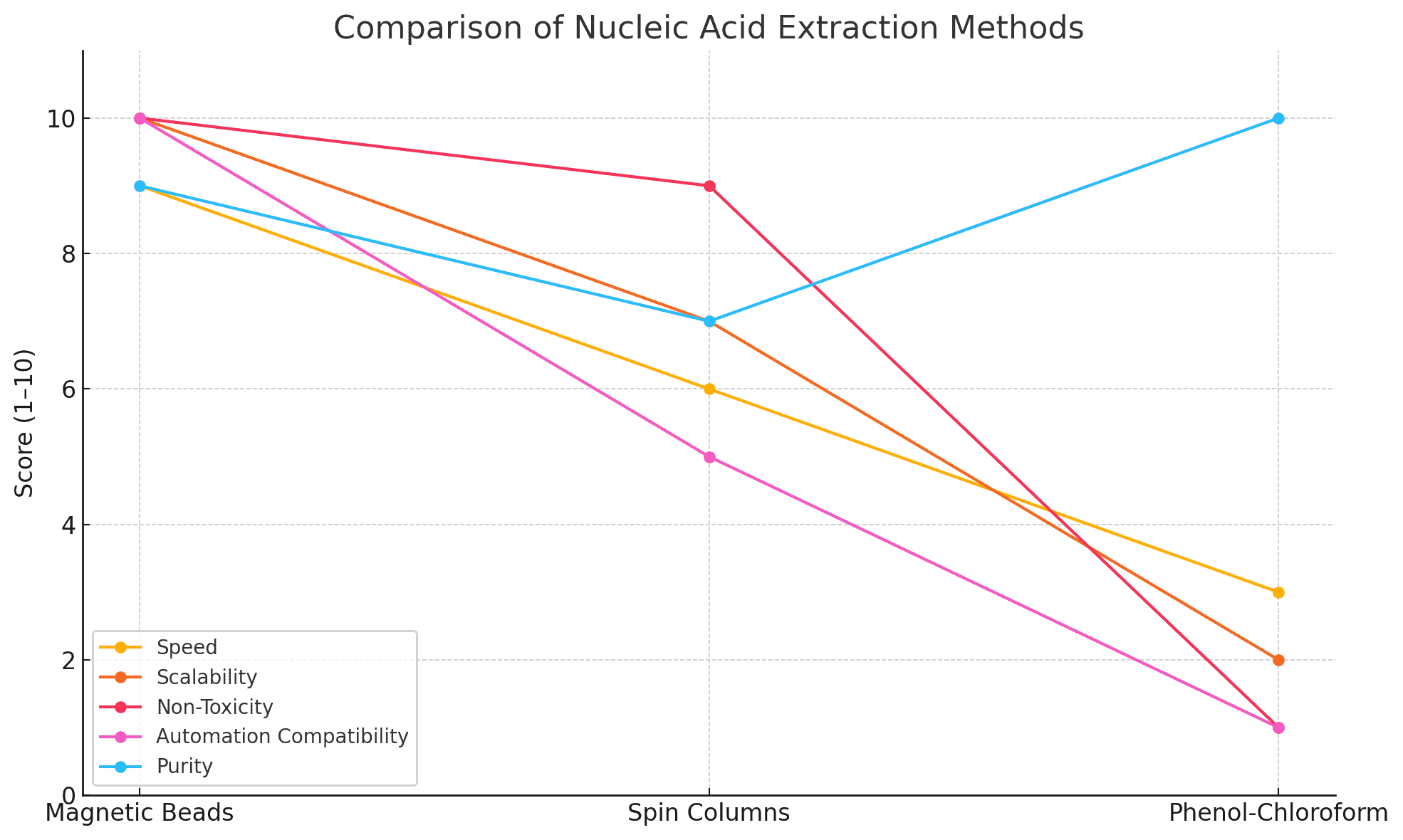
Magnetic bead-based nucleic acid extraction has become a cornerstone technique in molecular biology due to its scalability, reproducibility, and compatibility with automation. Magnetic beads offer a fast and efficient alternative to column- and chloroform-based methods, which often require manual centrifugation, hazardous chemicals, or limited input types.
The technology works by binding nucleic acids to magnetic particles under specific buffer conditions, allowing for rapid separation using a magnetic field. This technique is now widely used for isolating high-purity DNA and RNA from blood, tissue, swabs, saliva, microbial cultures, plants, and environmental samples.
For foundational references, visit:
Chemistry Behind Magnetic Bead-Nucleic Acid Binding
Magnetic beads are typically made from superparamagnetic iron oxide cores coated with a layer of silica or functional groups like carboxyl, amine, or streptavidin. The beads can bind DNA or RNA through hydrogen bonding, ionic interactions, or specific affinity mechanisms.
Chaotropic agents such as guanidinium hydrochloride or guanidinium thiocyanate disrupt water structure, helping expose the phosphate backbone of nucleic acids for bead interaction. Ethanol-based washes remove proteins, lipids, and salts. The final elution buffer (usually water or TE) is used to recover clean nucleic acids.
Detailed information is available at:
Step-by-Step Protocol for DNA and RNA Extraction
1. Sample Preparation
Whether using blood, cells, or plant tissue, the first step is proper homogenization. Tissue samples may require mechanical disruption or enzymatic digestion.
2. Lysis Buffer Composition
A lysis buffer typically contains:
-
Guanidinium salts
-
Detergents (SDS or Triton X-100)
-
Reducing agents (DTT or β-mercaptoethanol for RNA)
-
Optional: proteinase K
The goal is to release nucleic acids while preserving their integrity.
3. Binding to Beads
Add magnetic beads and a binding enhancer (e.g., isopropanol or ethanol). Mix thoroughly for 5–10 minutes to ensure complete interaction between nucleic acids and bead surfaces.
4. Magnetic Separation
Place the sample tube on a magnetic stand and allow beads to collect (usually ~2 minutes). Discard the supernatant without disturbing the pellet.
5. Washing
Typically involves two to three washes with 70–80% ethanol:
-
1st wash: removes salts
-
2nd wash: removes proteins and lipids
-
Optional 3rd wash: improves final purity
Spin briefly or air-dry beads before elution to avoid ethanol carryover.
6. Elution
Elute with nuclease-free water, TE buffer, or a low-salt solution. Warm the elution buffer to 65°C to improve yield, especially for RNA.
Performance Metrics and Quality Control
| Metric | DNA | RNA | Measurement Tool |
|---|---|---|---|
| A260/A280 ratio | 1.8–2.0 | 1.9–2.1 | Spectrophotometer (NIH Reference) |
| A260/A230 ratio | >1.8 | >1.8 | UV Absorbance |
| RIN score (RNA) | — | ≥7 | Bioanalyzer (NCBI) |
| Yield (ng/µL) | Sample-dependent | Sample-dependent | Nanodrop or Qubit Assay |
Automation and High-Throughput Compatibility
Magnetic bead systems are fully compatible with:
-
96-well plates
-
Liquid handling robots
-
Automated magnetic separators
Advantages:
-
Reproducible recovery
-
Minimal cross-contamination
-
Integration with PCR, RT-PCR, and sequencing
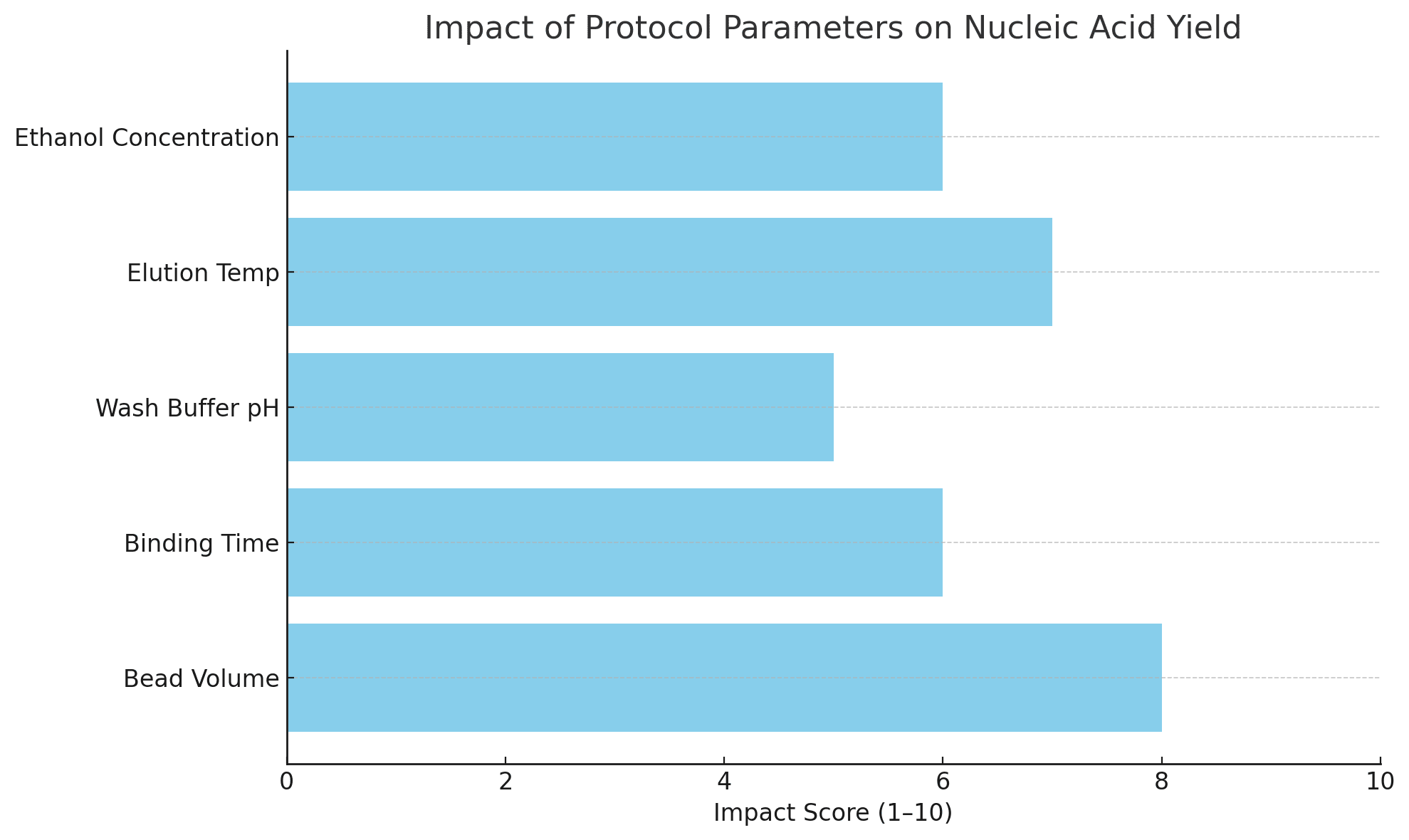
Tips for Optimizing Protocols
| Parameter | Optimization Strategy |
|---|---|
| Bead volume | Use 1.5–2x the nucleic acid content; avoid underloading or saturation |
| Binding time | Mix slowly for 5–10 minutes; vortexing may reduce efficiency |
| Wash buffer pH | Maintain neutral pH (7.0–7.5) for effective protein removal |
| Ethanol concentration | Ensure fresh ethanol at 70–80% to prevent salt precipitation |
| Elution volume | 30–50 µL is optimal for high-concentration elution |
For more optimization techniques:
Troubleshooting Low Recovery and Contamination
Low Yield
-
Check bead expiration or quality
-
Increase lysis time
-
Confirm proper magnetic separation
High Protein Contamination
-
Repeat ethanol washes
-
Extend drying time before elution
RNA Degradation
-
Use RNase-free tubes and filtered tips
-
Add RNase inhibitors during extraction
Helpful resources:
Applications of Magnetic Bead-Based Nucleic Acid Purification
-
PCR and RT-PCR: High-quality nucleic acids improve cycle thresholds
-
Next-Generation Sequencing (NGS): Cleaner input leads to more uniform coverage
-
Metagenomics: Effective for environmental or complex samples with inhibitors
-
Gene Expression Analysis: Reliable RNA integrity supports reproducibility
See also:
Conclusion
Magnetic bead-based extraction is a robust and reliable method that balances yield, purity, and scalability. With careful attention to buffer chemistry, magnetic bead properties, and protocol parameters, researchers can consistently obtain high-quality DNA and RNA suitable for a wide array of downstream applications.
This technique is especially suitable for laboratories requiring high-throughput, automation-ready workflows, and contamination-resistant nucleic acid recovery.
For more scientific guidance and validated protocols: