Site-directed mutagenesis (SDM) is a foundational technique in modern molecular biology that enables the intentional alteration of DNA sequences at precise locations. It has become a cornerstone in applications such as functional protein studies, enzyme engineering, plasmid development, and gene expression modification. The method covered in this guide focuses on PCR-based site-directed mutagenesis using high-fidelity enzymes, with a detailed breakdown suitable for academic, industrial, and synthetic biology laboratories.
This article is optimized for discoverability using strategic keywords such as “site-directed mutagenesis protocol,” “high-fidelity PCR,” “plasmid editing,” and “base editing workflow”. Numerous authoritative hyperlinks are included for further technical reference.
Introduction to Site-Directed Mutagenesis
Site-directed mutagenesis enables the introduction of a single base pair change, insertion, or deletion within a DNA sequence. The process typically involves the use of PCR primers carrying the desired mutation to amplify the entire plasmid, after which the methylated parental DNA is selectively degraded using DpnI endonuclease.
More on mutagenesis basics:
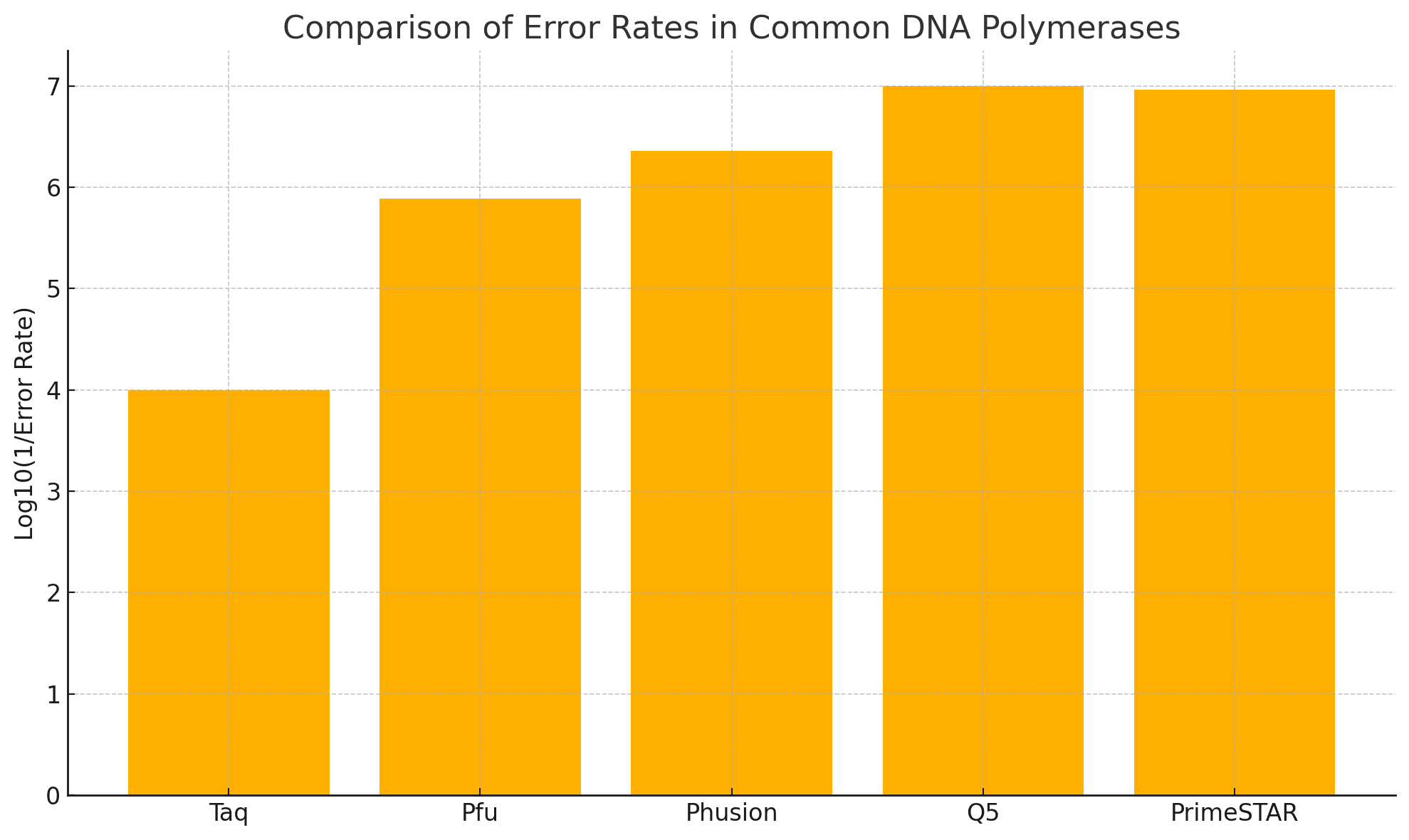
Why High-Fidelity DNA Polymerases Matter
High-fidelity DNA polymerases are crucial for accurate mutagenesis because they:
-
Possess proofreading activity (3’→5’ exonuclease),
-
Reduce PCR error rates to less than 1 in 1,000,000,
-
Enable amplification of long and GC-rich templates.
Recommended high-fidelity enzymes:
Validation studies:
Primer Design: The Heart of Mutagenesis
Each primer must:
-
Be 25–45 nucleotides in length,
-
Center the mutation within the sequence,
-
Possess 40–60% GC content,
-
Avoid secondary structures (hairpins, dimers).
Use these tools:
Reagents and Equipment Checklist
For thermocyclers and lab hardware:
Step-by-Step Protocol
1. PCR Reaction Setup
A 50 µL reaction typically includes:
| Reagent | Volume |
|---|---|
| High-Fidelity Buffer (5X) | 10 µL |
| dNTP Mix (10 mM each) | 1 µL |
| Forward Primer (10 µM) | 2.5 µL |
| Reverse Primer (10 µM) | 2.5 µL |
| Template DNA (5–10 ng) | 1 µL |
| DNA Polymerase | 0.5 µL |
| Nuclease-Free Water | 32.5 µL |
2. Thermal Cycling Conditions
Initial denaturation: 98°C, 30 sec
25 cycles of:
98°C, 10 sec
58–65°C, 20 sec
72°C, 30 sec/kb
Final extension: 72°C, 5 min
Hold: 4°C
PCR optimization guides:
3. DpnI Digestion
DpnI cleaves methylated (i.e., non-mutated parental) DNA:
-
Add 1 µL DpnI directly to PCR product.
-
Incubate at 37°C for 60 minutes.
DpnI protocol references:
4. Transformation
-
Use chemically competent E. coli cells.
-
Add 2–5 µL digested PCR product.
-
Ice incubation: 30 min
-
Heat shock: 42°C for 45 sec
-
Recovery in SOC medium: 1 hr at 37°C
-
Plate on selective antibiotic LB agar.
Transformation guide:
5. Colony Screening
-
Screen via colony PCR or plasmid isolation + sequencing.
-
Use Sanger sequencing for verification.
References:
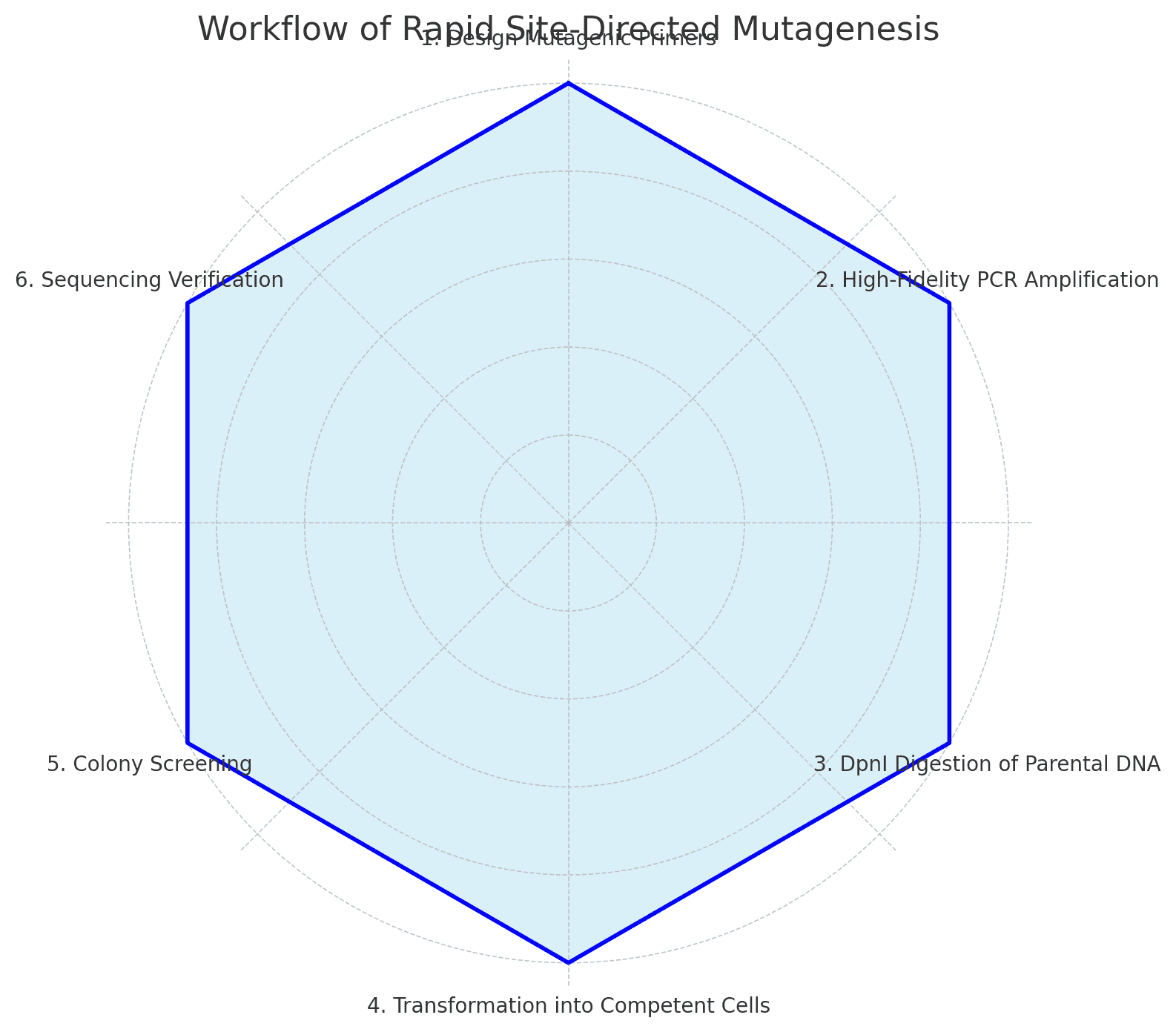
Chart: Workflow Summary
+----------------------+ +---------------------+
| Mutagenic Primers | --> | PCR Amplification |
+----------------------+ +---------------------+
|
v
+----------------------+ +---------------------+
| DpnI Digestion | --> | Competent Cell Txn |
+----------------------+ +---------------------+
|
v
+----------------------+ +---------------------+
| Colony Selection | --> | Plasmid Sequencing |
+----------------------+ +---------------------+
Common Issues and Fixes
| Issue | Suggested Fix |
|---|---|
| No colony growth | Redesign primers or check enzyme activity |
| Low PCR yield | Increase cycle number or adjust Mg²⁺ concentration |
| Unwanted mutations | Reduce PCR cycles; confirm polymerase fidelity |
| No mutation | Verify primer overlap and base mismatch location |
For troubleshooting:
Advanced Strategies
-
Multipoint Mutagenesis: Introduce multiple mutations simultaneously using overlapping primers.
-
Deletions and Insertions: Modify primer design to include desired deletions or inserted tags (e.g., FLAG, His6).
-
Degenerate Codon Mutagenesis: Introduce NNK codons for amino acid diversity in protein screening.
More resources:
Conclusion
PCR-based site-directed mutagenesis using high-fidelity DNA polymerases provides a fast, precise, and scalable method for targeted DNA manipulation. Its simplicity, cost-effectiveness, and reliability make it a staple protocol for any molecular biology lab. When performed with well-designed primers, optimized thermal cycling, and proper downstream handling, the protocol yields high success rates for diverse applications ranging from protein engineering to synthetic circuit design.