A Practical Guide for Accurate Nucleic Acid Quantification
Accurate and reliable quantification of nucleic acids is essential in modern molecular biology and genomic workflows. The Qubit™ Fluorometer from Thermo Fisher Scientific is a go-to platform for this purpose, offering enhanced specificity and sensitivity over spectrophotometric tools such as NanoDrop. However, the choice of Qubit assay kit—dsDNA, ssDNA, or RNA—must be informed by a deep understanding of your sample composition and experimental goals.
In this comprehensive guide, we will explore the core differences between these assay kits, common applications, technical limitations, and troubleshooting tips. We include direct references to verified technical resources from educational and governmental institutions for credibility and further study.
The Role of Fluorescent Quantification in Molecular Workflows
Traditional UV-based quantification at 260 nm measures total nucleic acid content, but cannot differentiate between DNA, RNA, and free nucleotides. In contrast, fluorescence-based Qubit assays use selective dyes that only bind to the target molecule—double-stranded DNA, single-stranded DNA, or RNA—to minimize overestimation.
According to NCBI, UV-based quantification can overestimate nucleic acid concentrations by up to 50% in samples with contaminants like phenol, guanidine, or proteins.
The Qubit platform is thus ideal for low-input samples and situations where high accuracy and specificity are required—such as NGS, RNA-seq, or digital PCR library preparation (CDC Lab Training).
Overview of Available Qubit Assay Kits
1. Qubit dsDNA Assay Kits
➤ Description:
The dsDNA kits are optimized for high-specificity quantification of intact double-stranded DNA. They are available in:
-
High Sensitivity (HS) for 0.2–100 ng samples
-
Broad Range (BR) for 2–1000 ng samples
➤ Best For:
-
Genomic DNA from extractions (UCSF Genomics Core)
-
DNA libraries for sequencing workflows
-
PCR amplicons and cloning vectors
➤ Technical Strength:
-
Does not bind to RNA or ssDNA
-
Minimal background from contaminants
➤ Limitations:
-
Cannot detect degraded DNA or single strands
-
Sensitive to pipetting variation and tube material (NIH Protocols)
2. Qubit ssDNA Assay Kit
➤ Description:
The ssDNA assay kit is designed for single-stranded DNA quantification, such as:
-
Oligonucleotides
-
Single-stranded viral genomes
-
Denatured PCR products
➤ Best For:
-
Synthetic ssDNA applications
-
Aptamer libraries (PubMed)
-
CRISPR single-strand donor templates
➤ Strengths:
-
High selectivity for ssDNA
-
Accurate detection of oligonucleotides
➤ Limitations:
-
May underperform with samples containing both ssDNA and dsDNA
-
Lower signal for structured ssDNA due to partial folding
3. Qubit RNA Assay Kits
➤ Description:
The Qubit RNA HS and BR kits are designed for quantification of total RNA in purified extracts.
➤ Best For:
-
Total RNA for transcriptome analysis (NIAID Genomics)
-
RNA quality screening before cDNA synthesis
-
Long non-coding RNA (lncRNA) and rRNA removal efficiency testing
➤ Strengths:
-
Highly selective for RNA
-
Resistant to DNA interference
➤ Limitations:
-
Not designed for small RNAs (miRNA, siRNA)
-
Requires RNase-free conditions (NCBI Bookshelf)
Real-World Examples and Use Cases
| Assay Kit | Example Application | Additional Tools Used |
|---|---|---|
| dsDNA HS | NGS library quantification | Illumina Prep Guide |
| ssDNA | Aptamer binding studies | PubChem Aptamer Data |
| RNA BR | RNA-seq transcript quantification | UCSC Genome Browser |
Protocol and Handling Considerations
To avoid variation and ensure precise results:
-
Use Qubit Assay Tubes only: Different plastics absorb light differently (FDA Laboratory Guidance)
-
Pipette carefully: Even a 1 µL error can significantly affect readings in low-volume protocols
-
Avoid contamination: RNase- or DNase-free reagents and barrier tips are essential
-
Equilibrate to room temperature: Cold reagents or samples can skew fluorescence readings
For reproducibility in high-throughput settings, ensure consistent vortexing and pipetting and use a single instrument when comparing samples.
Comparing Assay Specificity and Sensitivity
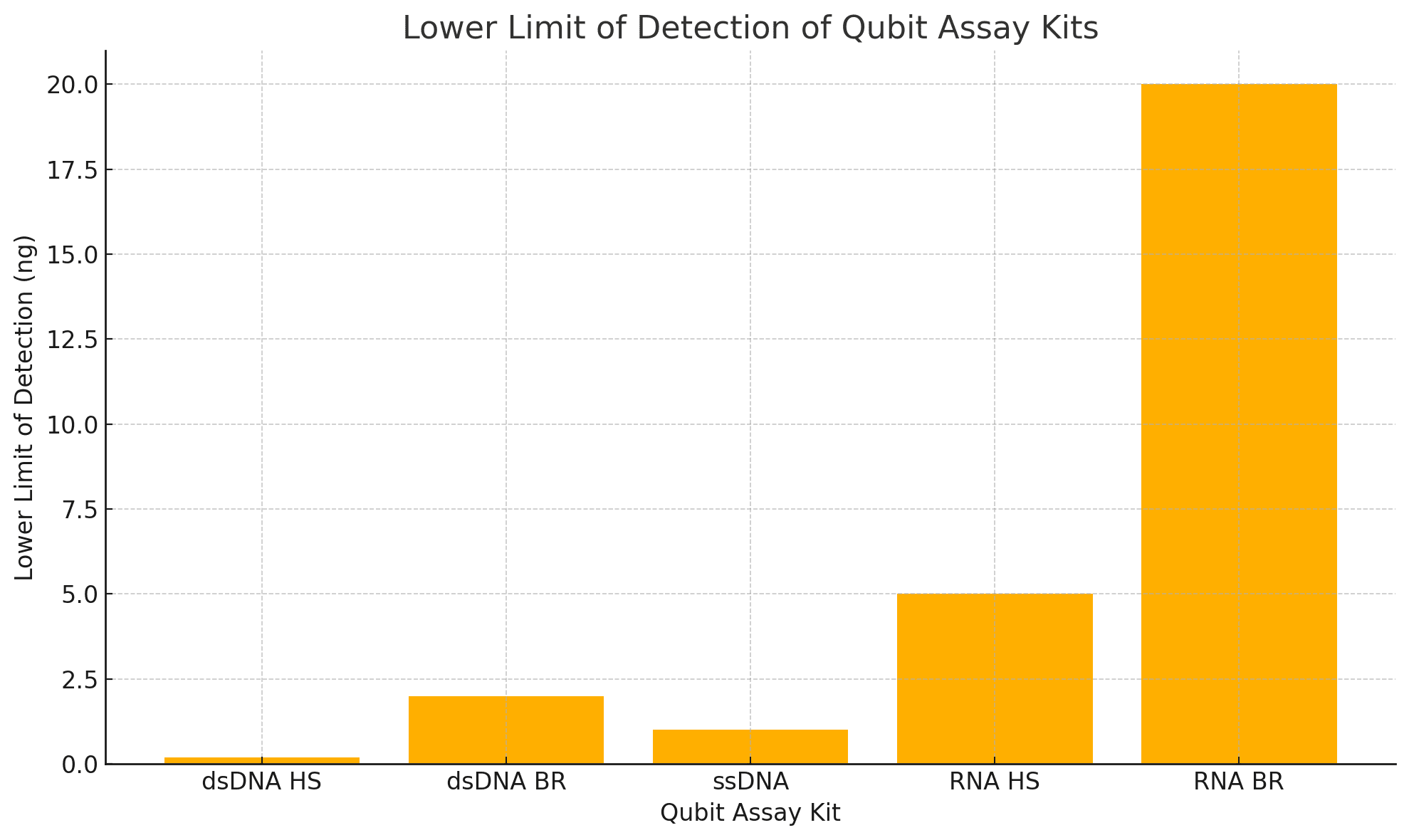
| Feature | dsDNA HS | dsDNA BR | ssDNA | RNA HS | RNA BR |
|---|---|---|---|---|---|
| Detection Range | 0.2–100 ng | 2–1000 ng | 1–200 ng | 5–100 ng | 20–1000 ng |
| DNA Specificity | ✔️ | ✔️ | ❌ | ❌ | ❌ |
| RNA Specificity | ❌ | ❌ | ❌ | ✔️ | ✔️ |
| ssDNA Specificity | ❌ | ❌ | ✔️ | ❌ | ❌ |
| RNase/DNase Safe | ✔️ | ✔️ | ✔️ | ✔️ | ✔️ |
Troubleshooting Common Problems
| Issue | Likely Cause | Recommended Fix |
|---|---|---|
| Low signal | Incomplete reagent mixing | Re-vortex dye and buffer thoroughly |
| Inconsistent replicates | Pipetting error | Use calibrated pipettes and slow draw/release |
| Background fluorescence | Residual DNA in RNA sample | Treat with DNase I before quantification |
| Fluorescence decay | Photobleaching or room light exposure | Read samples promptly after incubation |
For further technical validation, consult protocols from:
Integrating Qubit Results with Downstream Applications
A precise quantification step improves reproducibility in:
-
cDNA synthesis (NCBI Gene Expression Guide)
-
Normalization of NGS libraries (PubMed Central)
-
Preparation of DNA standards for qPCR (CDC Lab Quality)
Normalization using Qubit data is essential when pooling libraries for multiplexed sequencing (NCBI SRA).
Useful Educational Resources and Guides
Final Thoughts
Selecting the right Qubit assay kit is not just a technical choice—it is a foundational decision that impacts the accuracy and reproducibility of your entire experimental pipeline. Whether you’re quantifying intact genomic DNA, structured ssDNA, or total RNA, the specificity of Qubit kits ensures that only your molecule of interest is measured, free from overestimation caused by contaminants or degradation products.
By applying best practices and understanding the biochemical distinctions between dsDNA, ssDNA, and RNA, you can ensure successful downstream results, reduced variability, and higher data confidence.
For any molecular biologist, bioinformatician, or sequencing core facility, Qubit quantification remains a cornerstone of clean and consistent data output.